Faculty

Deepak Nair
ProfessorPhone : +91 80 2293 3535
E-Mail : deepak[at]iisc.ac.in
https://www.nanoorg.in/
Research Areas
Role of Dynamic Nanoscale Molecular Organization in the Signal processing at the Synapse
Research Details
Synapses, basic functional units of brain, are involved in the modulation of information flow between neurons. How synapses moderate this flow of information is key to functioning of the brain. Synapses control the transmission of information by differential recruitment, recycling and positioning of molecular components of the synapses. Our approach is to study the organization and dynamics of key synaptic molecules using a variety of interdisciplinary approaches. We aim to develop cutting edge microscopy and optogenetic tools to observe synapses at the nanoscale and real-time.
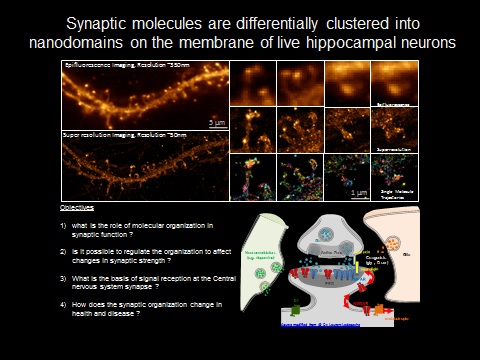
In the Central Nervous System (CNS), synapses form the basic functional units of connectivity between two neurons. The formation, remodeling and elimination of synapses refine the microcircuitry in the brain. Synapse is a complex molecular machine, which changes its structure and composition during neuronal development and plasticity. Synapse contains hundreds of proteins choreographed in to a micron sized machine overseeing the fidelity of brain function. The components of the synapses play a major role in synaptic transmission and synaptic plasticity, which are thought to underlie learning and memory. Interestingly most of the diseases has a direct impact on the number, position and trafficking of molecules in and out of synapse contributing towards synaptic loss or dysfunction thus affecting the normal behavior of the brain. However, it has been enigma how the information is processed at a single synapse by controlling function, position and regulation of several molecules.
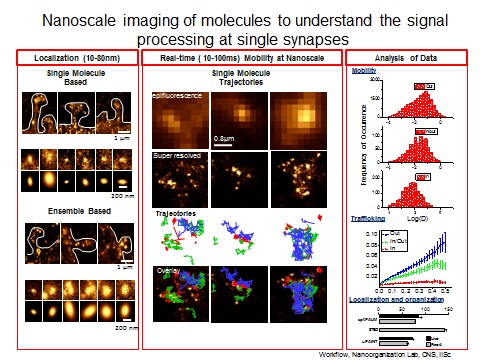
This is partly because of the inaccessibility to garner information to resolve structures less than few 100nm. The development of super-resolution imaging methods (Nobel Prize Chemistry 2014) that break the diffraction limit allows monitoring the synaptic organization at the nanoscale (10-50nm). Super resolution imaging provides evidences that synapse functions by local regulation and organization of nanometer sized protein domains.
The work in our lab attempt to dissect the fundamental role of dynamic nanoscale organization of synaptic molecules in how synapse process and relay information. To achieve this we follow an interdisciplinary research paradigm at the interface of high end microscopy, molecular biology and cellular neuroscience.
In order to comprehend the role of spatial heterogeneity in synapses, it is important to determine structural organization of molecular components of nano sized protein domains. Super-resolution imaging based on single molecule based techniques (STORM, PALM) will be used to observe the real-time organization of these sub-synaptic complexes in live cells at 10-50nm. The outcome is expected to provide a structural basis for nanoscale organization in synaptic transmission.
It has been shown that the synaptic morphology can affect synapse stability and function. How modifications in dendritic morphology direct nanoarchitecture of synapse will be vital in decoding the role of molecular organization and mobility in synaptic strength. Concurrently, how synapse decides that it needs more or less molecules in the postsynapse in response to presynaptic stimuli is still unclear. Optogenetic approaches, neurotransmitter uncaging, single molecule diffusion studies and super resolution imaging will be combined at the level of single synapses to address the role of mobility and localization in synaptic retention of molecules at resolution of few 10nms at millisecond timescale in live cells during synaptic plasticity.
Another important aspect is to compare the basic synaptic organization in transgenic mice models for neurodegenerative diseases to see how the spatio-temporal properties of the molecules are affected. In this case focus will be laid on the organization and recruitment of some of the key molecules implicated in these diseases. With all these information, we intend to create an in-silico model to demonstrate the role of nanoorganization of molecules in health and disease.